DNA fingerprinting has established itself as an efficient and highly accurate means of determining identities and relationships. It has practically revolutionized the field of forensics, especially concerning rape cases. DNA profiling, as the process is more appropriately called, involves the visualization of special segments of the human genome, which are unique to each individual. These special segments, called Standard Tandem Repeats (STR), can be cut out and separated from the rest of the DNA by two processes: mapping Restriction Fragment Length Polymorphisms (RFLP) and Polymerase Chain Reaction (PCR). After being separated from the other DNA, we can visualize these STR segments and separate them by size using gel electrophoresis. STR sequences can also be directly sequenced using DNA sequencing machines, but this method has yet to move into mainstream usage and is used primarily within the research community.
Introduction
It can exonerate a falsely accused person, while undeniably implicating a guilty party. It allows scientists to identify the remains of victims of highly disfiguring accidents or soldiers lost in battle. It can even settle child custody cases by identifying the child’s true biological parents. This amazing technology is DNA fingerprinting, and it is arguably the most powerful and accurate form of human identification currently used. DNA profiling, as it is more accurately called, is the process whereby we visualize a sample of DNA and determine its relationship to other DNA samples. This sounds simple in theory, but how we get from several people’s genomes-each of which contains over three billion elements-to a final conclusion is a complex process. There are currently two popular methods for DNA profiling, which use different elements of the DNA for identification: RFLP (Restriction Fragment Length Polymorphisms) and STR (Short Tandem Repeats). In this article we will examine the engineering and scientific basis behind these two powerful techniques as well as the future of identification by DNA profiling.
DNA: The Ultimate “How To” Guide
To understand the complicated technology behind DNA fingerprinting, we first need to understand some important background information about DNA structure and function. DNA molecules are tightly wrapped, like bundles of rope, into packages called chromosomes. Most humans have 23 chromosomes-collectively referred to as the human genome-which can be found in almost all of your living cells. Because almost every cell in your body contains a copy of your genome, it is very easy to sample, or in the case of criminals, leave behind copies of your DNA for profiling. DNA samples suitable for profiling have been retrieved from blood, urine, semen, saliva, hair roots, bone, and many other kinds of biological tissue [1].
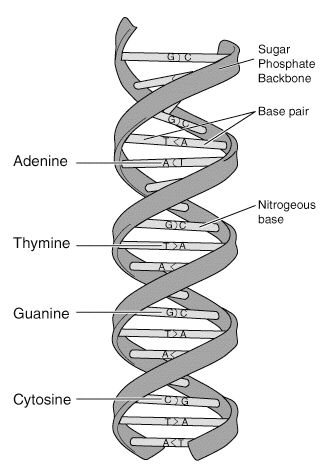
DNA molecules are very long and consist of two strands in a configuration resembling a zipper, except that the “teeth” of the two strands join head-to-head rather than interlock. The “teeth” of DNA molecules are called bases. There are four different biochemical bases, represented by four letters: adenine (A), thymine (T), guanine (G), and cytosine (C). These bases, arranged in a specific order along the two strands, are the letters of the genetic code (Fig. 1).
On a functional level, we can think of our DNA molecules as very long instruction manuals for making humans, with the bases (A, T, G, and C) as the words and sentences. The exception to this analogy is the presence of non-coding DNA in the human genome. Non-coding DNA does not contain instructions and, as far as today’s scientists can tell, serve no apparent function. An application of our analogy would look something like this: The foot bone’s connected to dknjbie andmnanofib sofbo ansoiyz boln the leg bone. These non-coding regions, though they do not serve any currently known biological purpose, are actually what make DNA profiling possible [2].
Another Use for DNA
Comparing several people’s genomes would, even with today’s best DNA sequencing machines, be a very difficult and time-consuming task if every single base of each person was examined. We are the same species after all, which means that over 99.9% of our genome is identical to every other human’s genome [3]. Looking for the unique 0.1% of three billion bases would be too much work for any practical application. Instead, in DNA profiling we look only at these enigmatic non-coding regions of DNA.
Many of the non-coding regions consist of repeated sequences called tandem repeats. For example, the tandem repeat “. . . AATGAATGAATGAATG . . .” is found on chromosome 11 of the human genome. Even though DNA sequences vary from person to person, the same Short Tandem Repeat (STR) sequences are found in similar locations in almost every human genome. What makes these sequences unique to each individual is the number of repeats. For instance, one person may have 25 repeats of AATG, while another has 111. By mapping multiple STR locations, we can create a unique “fingerprint” for any individual [4]. There will be other people with 111 AATG repeats, but it is unlikely that two people match in 100 or even eight other STR sequences [1]. Biotechnologists have devised several ingenious solutions in response to the problem of finding these special sequences on a molecule as complex as DNA.
Slice and Dice: Restriction Fragment Length Polymorphisms (RFLP)
DNA profile analysis was first made possible by the use of Restriction Fragment Length Polymorphisms (RFLP). Though it sounds complicated, RFLP is based on simple concepts. It is made possible by special ‘molecular scissors’ called restriction enzymes. Restriction enzymes are molecules found in bacteria, which cut DNA at specific sites. There are many different types of restriction enzymes and each type recognizes a different site on the DNA chain. For example, the enzyme TaqI cuts the DNA double strand everywhere it sees the sequence TCGA; BamHI cuts at GGATCC [2]. In bacteria these enzymes are present to destroy foreign DNA, such as might be present from an attacking virus. In the biological and biotechnological research community, restriction enzymes — which can now be ordered via catalogue — are used to perform a dizzying array of tasks, including RFLP DNA profiling.
In DNA profiling by RFLP analysis, scientists use specific restriction enzymes to cut STRs out of the DNA molecules. Scientists use restriction enzymes that cut roughly on either side of the STRs they are trying to find. In practice, usually two or three different restriction enzymes are used, so that enough sequences are isolated [5]. The final product of this stage is a test-tube of DNA fragments, which include both the desired STR sequences and the rest of the genome. The only thing left to do is isolate the STR sequences (restriction fragments) and determine how many times each sequence repeats (length polymorphisms). To accomplish this there is yet another technique, electrophoresis, which is detailed after the discussion of another method of obtaining STRs called Polymerase Chain Reaction.
The Copy Machine: Polymerase Chain Reaction (PCR)
The problem with RFLP analysis is that it requires a relatively large amount of DNA, which in turn requires a relatively large amount of sample. This hinders its usefulness in the field of forensics. By contrast, the Polymerase Chain Reaction allows us to develop a DNA profile from as little DNA as might be deposited by licking an envelope [3]! PCR can do this because instead of cutting out the target STR sequences from the rest of the genome, it amplifies the sequences by replicating them over and over. DNA is replicated by the naturally occurring enzyme DNA polymerase. In nature, the two strands of the DNA molecule are replicated individually and in their entirety — there is never replication of just one section of DNA. To force the replication of specific sequences of the genome, scientists have developed special DNA primers that recognize and stick to specific sequences, and enable DNA polymerase to replicate only those sequences enclosed by the primers. Since we know the desired STR sequences, and we know some of the bases before and after the STR, we can construct primers that will attach themselves on either side and allow replication of the sequence in-between.
PCR is a cyclic process that, as long as the temperature is controlled, is a self-perpetuating “chain reaction.” First, the DNA sample is heated and the primers are added. The sample is then cooled, and the primers attach themselves to the DNA. Then a special heat-resistant DNA polymerase called Taq polymerase is added and immediately begins to replicate the STR sequences. Now for every original DNA molecule in our sample, we have two copies of each desired STR sequence. We can repeat this process many times by fluctuating the temperature, each cycle doubling the amount of STR sequence fragments. The final product of this stage is a test-tube containing a high concentration of our target STR sequences [2].
Gel, Electricity, and Dye: Measuring DNA Fragments
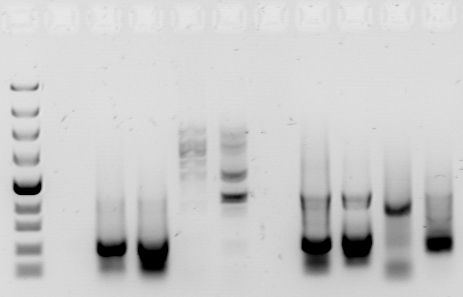
We now have several individuals’ DNA profiles in the form of fragmented DNA in separate test tubes. How can we measure these microscopic fragments? The answer is gel electrophoresis — an ingenious method of visualizing DNA fragments and separating them by size.
Gel electrophoresis exploits one of the properties of the DNA molecule — its negative charge. In electrophoresis, we inject a few microliters of a DNA sample into tiny wells at one end of a rectangular gel as seen. On the same end of the gel we place a negative electrode; on the opposite end, we place a positive electrode. When we turn on the electrodes, the negatively charged DNA fragments are pulled across the gel by the positive electrode. But because the gel is thick-about the consistency of very solid Jell-O-the fragments do not move very quicklyAs you might expect, the shorter DNA fragments, because of their smaller size, travel faster than longer DNA fragments. After a certain amount of time, the DNA fragments are distributed by size (length) along the gel, with the smallest fragments near the positive electrode and the larger fragments near the negative electrode [2].
Once the gel has been “run,” the only task remaining is to illuminate the DNA fragments. With PCR, the entire gel is simply soaked in ethidium bromide, a staining solution. The DNA fragments appear as little bands on the gel and we can determine their approximate length by their position on the gel (Fig 2). RFLP is a bit more difficult because whereas PCR isolates the target sequences from the rest of the genome, the product of RFLP is a conglomeration of DNA fragments, most of which are not needed. To illuminate the DNA profile from an RFLP analysis, we first need to expose it to radioactively labeled probes, which specifically target the STR sequences that we are trying to find. Then we can expose it to an X-ray film, which will visualize only the fragments that were detected by the radioactively labeled probes. The final picture will look very similar to the PCR DNA profile [2].
As long as the same techniques are used in preparing all the samples in a test, a direct comparison of the DNA profiles is an accurate way to match identities or confirm relationships. A person’s DNA profile will be approximately a 50/50 combination of his or her mother and father [3]. An exact DNA profile match either indicates that both samples were taken from the same person, or from that person’s identical twin [5].
A Look to the Future
The motivating idea behind these two techniques is to examine certain areas of the genome since analyzing the entire genome base-for-base would be too complicated and time-consuming. Nevertheless, DNA sequencers are being used with PCR in research [6] and the Armed Forces Institute of Pathology [1]. As DNA sequencing technology becomes more efficient and more affordable, it is likely that we will see an increase in the use of DNA sequencers to provide a more detailed, though not necessarily more correct, DNA profile. Could we eventually be able to use our DNA profile — maybe from a tiny sample taken from the tip of our finger — like a fingerprint or signature? Probably not in the foreseeable future, but as our technology grows along with knowledge of our DNA, we can anticipate increased usage of DNA profiles as a means of identification throughout our lifetime.