Abstract
DNA has been introduced to the computer science field as one of the newest materials used for computer construction and computational mechanics. Its unique chemical properties make it faster and smaller than traditional computers– able to perform parallel operations on enormous amounts of data. Since Leonard Adleman’s first experiment with this media, others have created logic gates and cellular automata with the hopes of creating universal computers. Though DNA may not entirely replace traditional computers, recent innovation in DNA will transform computer mechanics.
Introduction
When asked to think of a computer, most people envision some variation of an electronic device, which uses resistors, electron currents, semiconductors and an exponential list of “techy” gadgets. Though popular and extremely efficient, electronics are not the only components that can make up a computer. Moving beyond gears, electromechanical relays, and light, engineers have begun to venture into the world of quantum computers. Within this field, DNA, whose strands and chemical properties allow for unique computing methods and applications, stands out as a promising media for future computational mechanics.
What Is DNA Computing?
Before considering alternate methods of computation, we must first let go of the notion that all computers need to work in binary or use electricity. Instead, we must focus on the fundamental nature of computation. At its most basic level, a computer is anything that can receive an input and reliably produce a desired output. This input and output can take any form– from electric signals to a configuration of beads on a string. Take, for example, a measuring cup. Though unconventional, this simple device can fit under the broad definition of a “computer”. The input is “x” amount of water, and the output is defined number (marked on the outer face of the cup) that quantifies exactly how much water was poured in. Of course, this kind of computer can only solve one problem, but this example is valuable since it highlights the simple, physical properties that give it computational character.
DNA is another example of an unconventional computer. Like physical properties of silicon ease computation in traditional computers, physical properties of DNA influence DNA computing. DNA computing uses DNA structures to transform some input into an output and solve a computational problem. This is not to be confused with using computers to analyze DNA in efforts like the Human Genome Project. For this article, the DNA is the computer, and whichever problem needs to be solved must first be encoded in such a way that the DNA is able to read and analyze incoming data in a consistent, coherent manner. The resulting output can be anything from a daughter strand of DNA to an engineered chemical property of a molecule to a cellular reaction.
Why DNA?
There are many advantages to using DNA over traditional circuits, especially considering DNA’s nanoscale size and high information density. DNA needs only one cubic nanometer per bit of information, whereas traditional computers may need 1012 cubic nanometers [1]! Since chemical reactions (or “operations”) happen almost simultaneously instead of consecutively, like in a traditional computer, a DNA computer can perform many processes in parallel, significantly increasing speed of computation. A small experiment could work at 100 Teraflops (1012 operations per second), which approaches near supercomputer speeds [1]. In addition to its computational properties, DNA is abundant, and can be folded into different shapes. Its inherent chemical properties allow it to react directly to environmental changes while a traditional computer would need a specific scanner replicate of those changes.
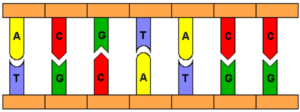
DNA shares many similarities with current computing practices. “Information” is encoded in DNA as four bases, Adenine (A), Thymine (T), Guanine (G), and Cytosine (C) (as depicted in Figure 1), which mirrors the binary form of storage in electrical computers [2]. Since the original purpose of DNA is to store data, the genetic code of an organism, it is only a small step to reprogram it store alternative data of interest. The bases’ natural pairing properties are also useful for computation. A binds to T and G binds to C, meaning that complementary sequences of these bases will bind together.
History
The first DNA computing experiment was conducted in 1994 by Leonard Adleman, professor of Computer Science and Molecular Biology at the University of Southern California. His experiment was created to solve a computational problem called the Hamiltonian Path Problem that requires determining whether a path exists in a group of connected points that goes through each point exactly once [3]. He chose this problem because despite its simplicity, it requires a long time to compute. The greater purpose of the experiment was not to solve the problem but expose the computational ability of DNA.
He first encoded the problem into DNA as sequences of bases A, T, G, and C. By manipulating and analyzing the DNA, he was able to reach the solution. When mixing these sequences of DNA, complementary sequences would bind together, ultimately producing all possible paths as DNA strands [3]. By performing chemical reactions and sorting the DNA by length, he narrowed the possible solutions down until it was shown that either no correct solution exists or the correct solution is the only strand left. At the end of Adleman’s procedure, a strand remained which displayed the solution: the correct path through the graph. The whole procedure took 7 days for a collection of 7 points, but it proved that DNA is a feasible medium for computation [3].
Further Development
Though Adleman’s experiment proved that DNA could be used for computation, it only showed this for a single problem. However, this experiment was the first step in developing a universal computer—one capable of solving any problem that can be solved on a traditional computer.
One step toward this was creating boolean logic gates. A logic gate functions by receiving truth values as input (encoded as 1 or 0, true or false) and outputting a truth value based on these inputs. A traditional computer contains logic gates such as an AND gate which outputs true if both conditions input are true, or an OR gate, which outputs true if either condition is true. By stringing these gates together, one can create various operations, and, subsequently, a fully working computer.
Okamoto, Tanaka, and Saiko from Kyoto University found a method to create these logic gates, allowing for more universal DNA computation [4]. One strand would serve as the logic gate and would be paired with an “input” strand. Electron holes can travel along DNA strands and be measured, serving as the output. By combining different base pairs, one can affect how these holes are transported along the strand to create the logic gates required for computation.
Applications
A long-term advantage to DNA computing is the miniaturization of computing. Despite recent advances in this field, however, reading and understanding the results of a DNA computation is still too difficult to justify its use over current computers. Much like quantum computers, DNA is very specialized and may never truly replace the electronic computer. For very specific jobs, however, it may be the optimal tool for the job. By manipulating base pairs, DNA is able to self-assemble into structures that can hold molecules or even perform computation in the folding itself [5]. Because the structures assemble with predictable patterns, one can produce a cellular-automaton, a rule based system that can be used to simulate a full computer. By manipulating the rules, one can build specific structures or even “programs” [5].
The most exciting prospect for DNA computing is self-diagnosing medicine. In some cases like the treatment of leukemia or lymphoma, treatment given can be harmful to cells if applied in the wrong places. Chemotherapy can be an intrusive and painful procedure for patients; DNA computing could be used to mitigate the harmful effects. The process is relatively simple: treatment is only released if a series of yes-or-no conditions are met, so a molecule can “diagnose” based on these preset conditions [6]. By only seeking proteins found in cancer cells, a DNA computer module can detect cancer cells and release a cytotoxin only if it encounters one, preventing unnecessary damage to non-cancerous cells in the body. The most impressive part of this self-diagnosing medicine is its flexibility, since not only the treatment released can be varied, but also the conditions for treatment and the degree of certainty necessary for a positive diagnosis [6].
Why It’s Important
It is unlikely that the smartphones and supercomputers of the future will be running on DNA. For many problems, the amount of DNA needed for solutions grows exponentially, requiring bathtubs full of DNA just to compute. Consequently, universal DNA computers are difficult to efficiently maintain. The future of this field relies on discovery of a way to put DNA’s unique properties to good use. Through creative use of this new computing power, we could treat many diseases more elegantly, and even do human body engineering on a cellular level [1]. DNA computing marks a step towards much smaller, more complex nanotechnology.
As a medium for computation, DNA is still in its infancy. Just like the electric computer when it was first developed, DNA has a lot of room for growth. By expanding beyond traditional ideas of a computer, researchers have discovered a new field of study and an impressive new method for computing.
References
[1] J. Parker, “Computing with DNA,” EMBO reports, vol. 4, no. 1, pp. 7–10, Jan. 2003.
[2] M. Amos, “DNA computing,” in Encyclopædia Britannica, Encyclopædia Britannica, 2016. [Online]. Available: https://www.britannica.com/technology/DNA-computing. Accessed: Sep. 15, 2016.
[3] L. Adleman, “Molecular computation of solutions to combinatorial problems,”Science, vol. 266, no. 5187, pp. 1021–1024, Nov. 1994.
[4] A. Okamoto, K. Tanaka, and I. Saito, “DNA logic gates,” Journal of the American Chemical Society, vol. 126, no. 30, pp. 9458–9463, Aug. 2004.
[5] P. W. K. Rothemund, N. Papadakis, and E. Winfree, “Algorithmic self-assembly of DNA Sierpinski triangles,” PLoS Biology, vol. 2, no. 12, p. e424, Dec. 2004.
[6] E. Shapiro and Y. Benenson, “Bringing DNA computers to life,” Scientific American sp, vol. 17, no. 3, pp. 40–47, Sep. 2007.
[7] Brain, M. “How Cells Work,” How Stuff Works. Access: http://science.howstuffworks.com/life/cellular-microscopic/cell6.htm




